Computational Evolution of Anti-PD‑1 Antibodies Induces Structural Refolding for High-Affinity Interactions #468
Authors
Yuanjun Shi, Yeil Kim, Pulan Liu, Jimin Wang, Shaogeng Tang, Victor S. Batista
Abstract
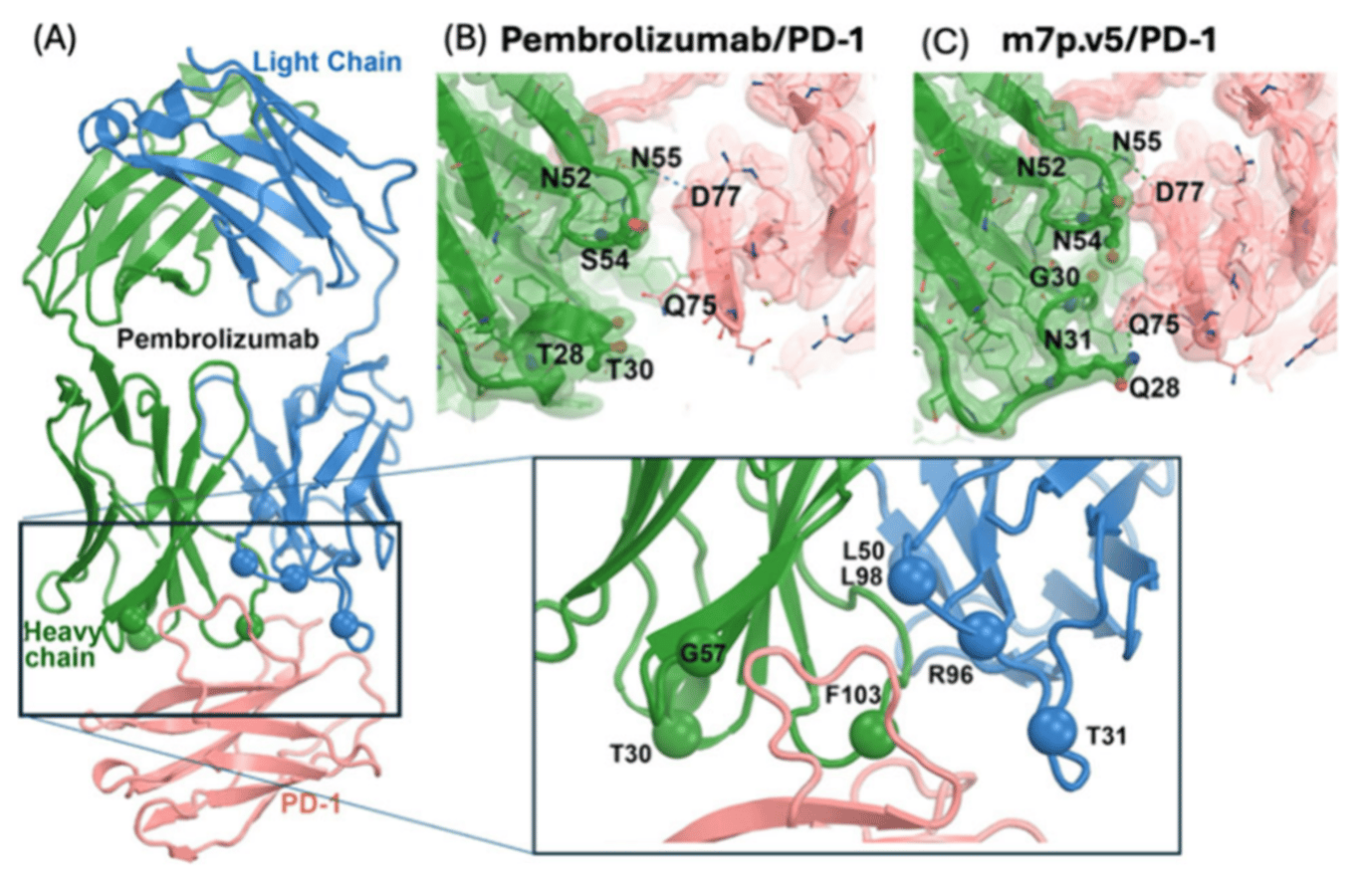
Checkpoint inhibitors targeting the PD-1/PD-L1 axis are key immunotherapies, but the dynamic and flexible nature of PD-1 complicates rational antibody engineering. Here, we use computational saturation mutagenesis, AlphaFold prediction, and molecular dynamics (MD) simulations to evolve pembrolizumab variants with suitable binding. Seven engineered antibodies form additional salt bridges and hydrophobic contacts via refolding of both the antibody and the PD-1 interface. One variant, m7p.5, displays improved biphasic kinetics and high-affinity binding (K_D,apparent = 62 pM). Structural changes include an α-helix to loop transition in the antibody heavy chain and a 4.6-Å Cα shift of a PD-1 loop. These results show that computational evolution can access binding modes inaccessible to traditional rigid structural design, enabling high-affinity antibodies for flexible targets. It is demonstrated that our integrated computational approaches including MD simulations can generate new picomolar high-affinity antibodies targeting specific epitopes of proteins that may be intrinsically flexible and are difficult to target with reasonable computational cost, which would be far less than an experimental cost for finding new antibodies with equivalent binding affinities. This study provides a new tool that can be combined with other artificial-intelligence-based antibody generation against PD-1 from the existing anti-PD-1 antibody library with broad applications in protein-protein interactions.